Robust autonomous visual detection and tracking of moving targets in UAV imagery
The use of Unmanned Aerial Vehicles (UAVs) for reconnaissance and surveillance applications has been steadily growing over the past few years. The operations of such largely autonomous systems rely primarily on the automatic detection and tracking of targets of interest. This paper presents a novel automatic multiple moving target detection and tracking framework that executes in real-time and is suitable for UAV imagery. The framework is based on image feature processing and projective geometry and is carried out on the following stages. First, outlier image features are computed with least

Scale-adaptive object tracking with diverse ensembles
Tracking by detection techniques have recently been gaining increased attention in visual object tracking due to their promising results in applications such as robotics, surveillance, traffic monitoring, to name a few. These techniques often employ semi-supervised appearance model where a set of samples are continuously extracted around the object to train a discriminant classifier between the object and the background whereas real-time performance is attained by using reduced object representations as in the case of the compressive tracking algorithm. However, because they rely on self
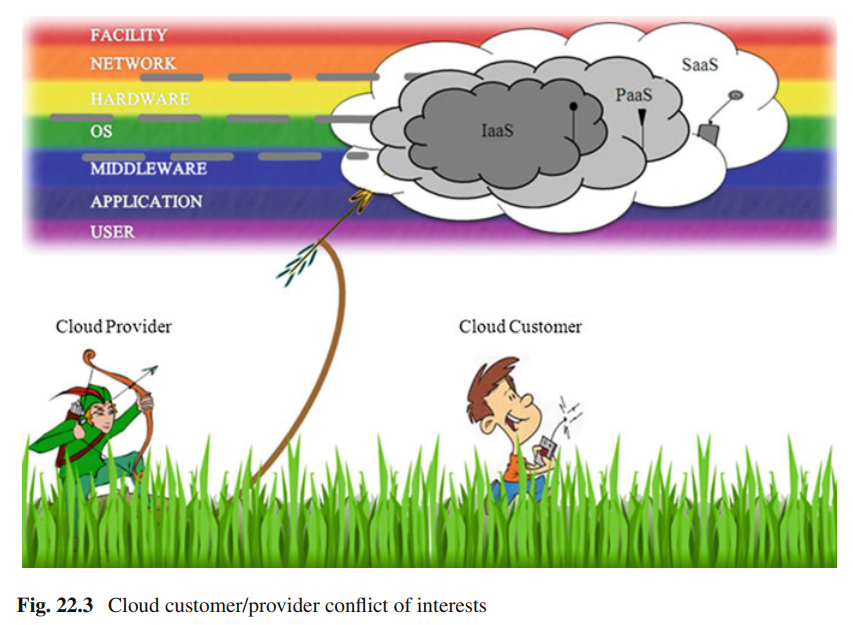
Towards cloud customers self-monitoring and availability-monitoring
As an attractive IT environment, Cloud Computing represents a good enough paradigm which governments, national entities, small/medium/large organizations and companies want to migrate to. In fact, outsourcing IT related services to Cloud technology, needs monitoring and controlling mechanisms. However, Cloud Customers cannot fully rely on the Cloud Providers measurements, reports and figures. In this book chapter, we cover the two Cloud Computing operation sides. For the first operation side, we provide advices and guidelines for Cloud layers which can be under Cloud Customer control, to allow
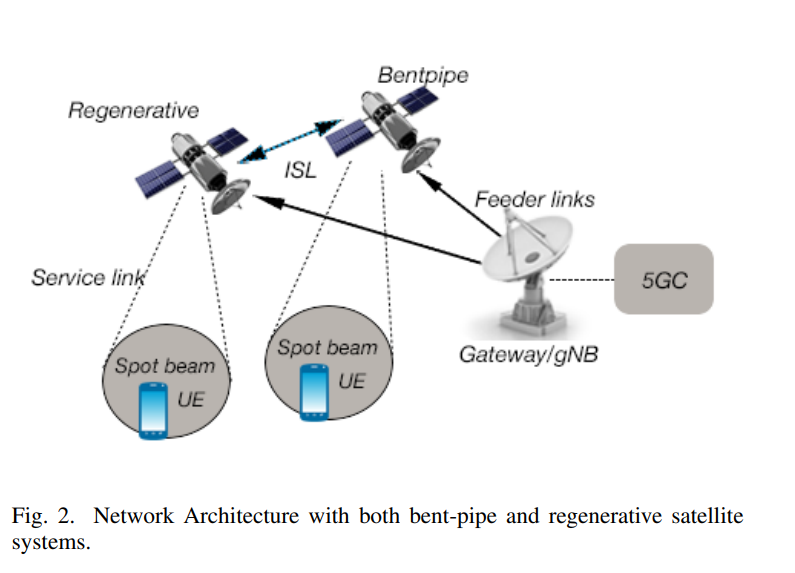
5G and Satellite Network Convergence: Survey for Opportunities, Challenges and Enabler Technologies
Development of 5G system as a global telecommunication infrastructure is accelerating to realize the concept of a unified network infrastructure incorporating all access technologies. The potential of Low Earth Orbit (LEO) constellation systems has emerged to support wide range of services. This could help to achieve 5G key service requirements for enhanced Mobile Broadband (eMBB), Massive Machine-Type Communications (mMTC), and Ultra-Reliable Low-Latency Communication (URLLC). The integration of satellite communications with the 5G New Radio (NR) is stimulated by technology advancement to

Vision capabilities for a humanoid robot tutoring biology
Robots are expected to be the future solution in various fields. One of these fields is education. Teachers, students and robots have to work together to make this assumption true. For this, robots must have the adequate capabilities that can help them succeed. Vision of the robot is an essential tool that the robot uses to perform several tasks. Hence, it has to be taken into consideration, the steps and the expectations for this robot vision system to surpass at least the very basic skills. Detection, recognition, and localization are the basic skills that we are implementing in our
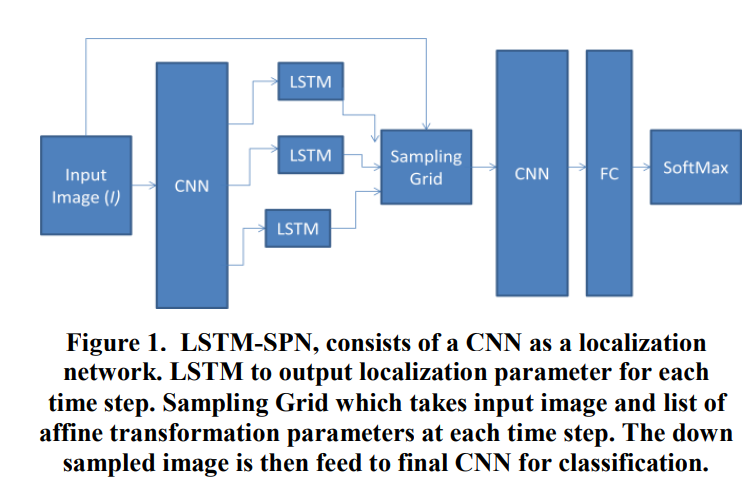
Optical character recognition using deep recurrent attention model
We address the problem of recognizing multi-digit numbers in optical character images. Classical approaches to solve this problem include separate localization, segmentation and recognition steps. In this paper, an integrated approach to multi-digit recognition from raw pixels to ultimate multi class labeling is proposed by using recurrent attention model based on a spatial transformer model equipped with LSTM to localize digits individually and a subsequent deep convolutional neural network for actual recognition. The proposed method is evaluated on the publicly available SVHN dataset where
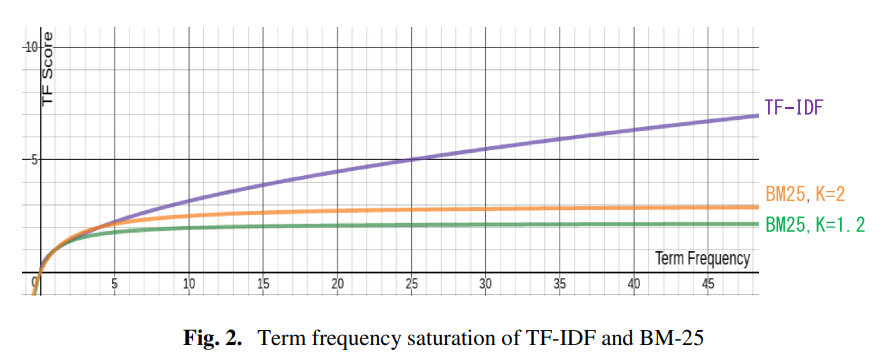
Behaviorally-Based Textual Similarity Engine for Matching Job-Seekers with Jobs
Understanding both of job-seekers and employers behavior in addition to analyzing the text of job-seekers and job profiles are two important missions for the e-recruitment industry. They are important tasks for matching job-seekers with jobs to find the top relevant suggestions for each job-seeker. Recommender systems, information retrieval and text mining are originally targeted to assist users and provide them with useful information, which makes human-computer interaction plays a fundamental role in the users’ acceptance of the produced suggestions. We introduce our intelligent framework to
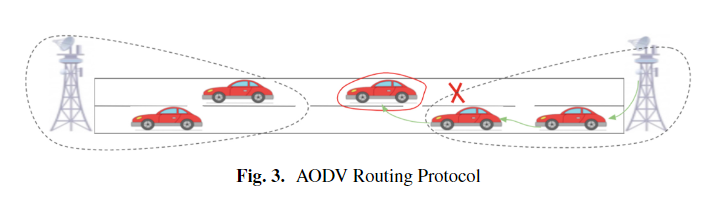
A Comprehensive Survey on Vehicular Ad Hoc Networks (VANET)
Vehicular Ad Hoc Networks is an evolving research field that has the potential to address safety on roads. This tends to attract car manufacturers and suppliers to develop in evolving the industry vision. VANET demonstrates a different kind of communications targeting the main objective, which is safety besides entertainment services. This is achieved using the Internet through the infrastructure units located aside on the roads. In this paper, the main concern is safety-oriented communication, V2V, and V2I, which are often used together to achieve the required safety objectives. We focus on

Efficient Quering Blockchain Applications
Industrial blockchain applications have recently risen to the top of the scientific and industrial communities' priority lists. This is due to their practical capabilities in resolving many issues in various industrial domains. Visibility and traceability to a large volume of trusted data benefit members of a consortium of industrial companies and associated organizations. Data scientists who query data and use statistics and Machine Learning to solve a wide range of interesting problems benefit as well. This paper introduces a blockchain application Query Engine based on upgradable Smart
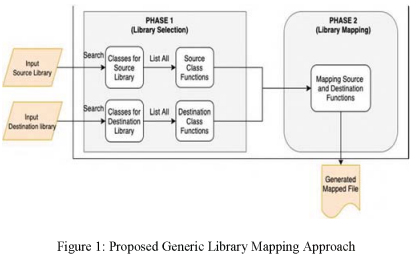
Generic Library Mapping Approach for Trans-Compilation
Cross-platform mobile development is a widely used framework due to its nature of building an app using one development life cycle and deploying it to multiple platforms like Android and iOS. Many cross-platform solutions were recently developed to convert from one platform to another using Trans-compilation approach as Trans-Compiler Android to IOS Conversion (TCAIOSC) and Trans-Compiler Based Mobile Applications code converter. The limitations of these solutions is that they are unable to convert a library function that was not previously included by the developer. In this paper, a generic
Pagination
- Previous page ‹‹
- Page 26
- Next page ››