Simple implementations of fractional-order driving-point impedances: Application to biological tissue models
A novel procedure for the circuit implementation of the driving-point impedance of frequency-domain material models, constructed from fractional-order elements of arbitrary type and order, is introduced in this work. Following this newly introduced concept, instead of emulating separately each fractional-order element in the model under consideration, the direct emulation of the complete model can be achieved through the approximation of the total impedance function. The magnitude and phase frequency responses of the impedance function are first extracted and approximated through curve-fitting

Response Surface Methodology Optimization of Mono-dispersed MgO Nanoparticles Fabricated by Ultrasonic-Assisted Sol–Gel Method for Outstanding Antimicrobial and Antibiofilm Activities
Magnesium oxide (MgO) nanoparticles are one of the highly significant compounds in construction. The novelty concentrated on using sol–gel technique coupled with ultrasonication for synthesis of MgO nanoparticles to prevent the agglomeration and its effect on the size was investigated. The synthesized samples were characterized by TGA, DSC, XRD, FTIR, SEM, EDX mapping, DLS, and HRTEM. Antimicrobial and antibiofilm activities of MgO nanoparticles were investigated against multidrug-resistant microbes causing-urinary tract infection (UTI). TGA, XRD, and FTIR characterization were used to
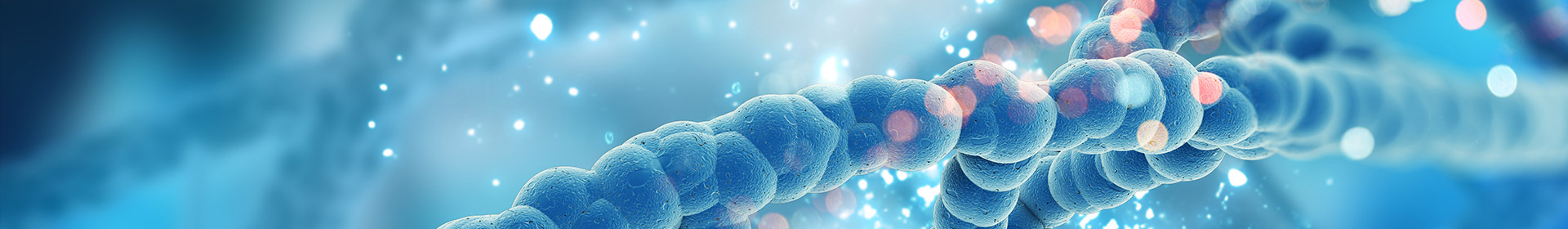
Gelatin Loaded Titanium Dioxide and Silver Oxide Nanoparticles: Implication for Skin Tissue Regeneration
Treatment of burn wounds has many requirements to ensure wound closure with healthy tissue, increased vascularization, guarantee edema resolution, and control bacterial infection. We propose that titanium oxide (TiO2) nanoparticles (NPs) will be more efficient than silver dioxide (Ag2O) in the treatment of burn wounds. Herein, gelatin loaded NPs (GLT-NPs) were evaluated for their efficacy to regenerate second-degree burn wound in rabbit skin. TEM results revealed that the average particle sizes were ⁓ 7.5 and 17 nm for Ag2O and TiO2 NPs, respectively. The results of the in vivo application of
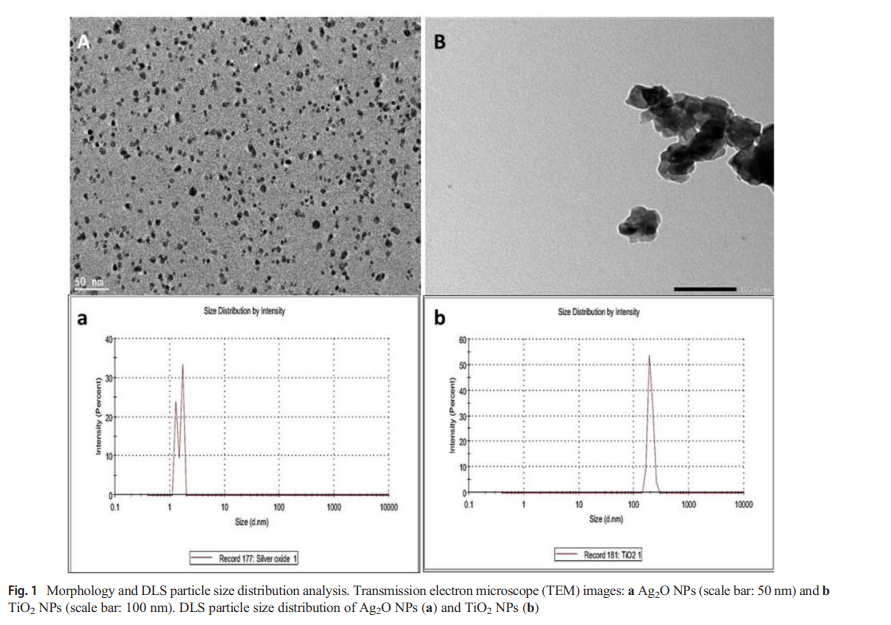
Therapeutic and diagnostic potential of nanomaterials for enhanced biomedical applications
Biomedical applications of nanomaterials have received considerable attention and interest from many researchers over the past decade due to the key role they can play in enhancing public health. Different types of nanomaterials possess both diagnostic and therapeutic potential owing to their outstanding properties compared to their bulk counterparts. Herein, we present, analyze and provide significant insights and recent advances about the promising biomedical applications of nanoparticles including bioimaging of biological environments and its role as a significant tool for early detection
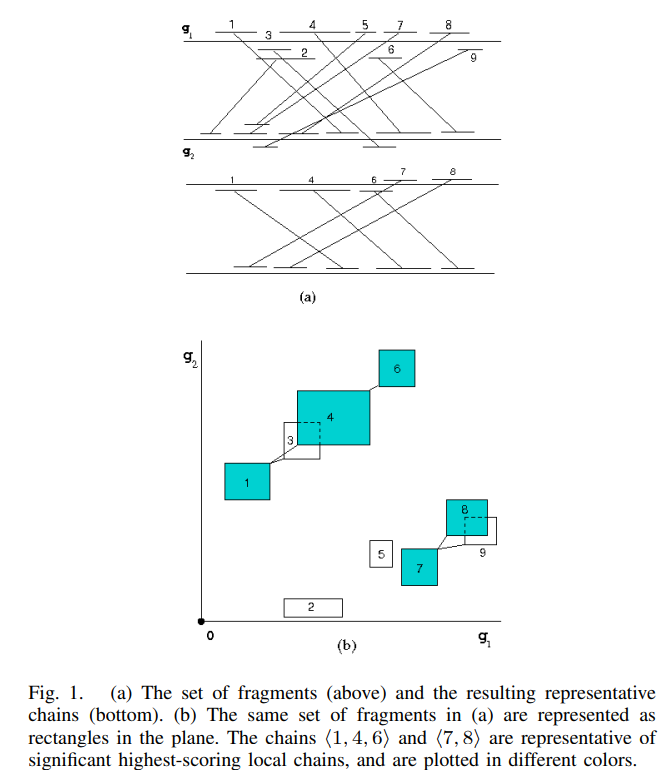
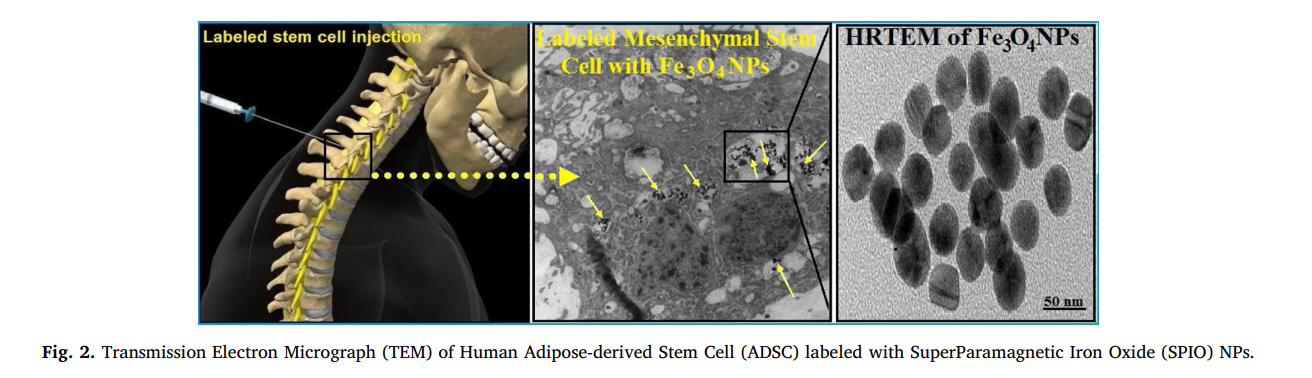
VisCHAINER: Visualizing genome comparison
Visualization of genome comparison data is valuable for identifying genomic structural variations and determining evolutionary events. Although there are many software tools with varying degrees of sophistication for displaying such comparisons, there is no tool for displaying dot plots of multiple genome comparisons. The dot plot mode of visualization is more appropriate and convenient than the traditional linear mode, particularly for detecting large scale genome deletions, duplications, and rearrangements. In this paper, we present VisCHAINER, which addresses this limitation, and displays
Study of Energy Harvesters for Wearable Devices
Energy harvesting was and still an important point of research. Batteries have been utilized for a long time, but they are now not compatible with the downsizing of technology. Also, their need to be recharged and changed periodically is not very desirable, therefore over the years energy harvesting from the environment and the human body have been investigated. Three energy harvesting methods which are the Piezoelectric energy harvesters, the Enzymatic Biofuel cells, and Triboelectric nanogenerators (TENGs) are being discussed in the paper. Although Biofuel cells have been investigated for a
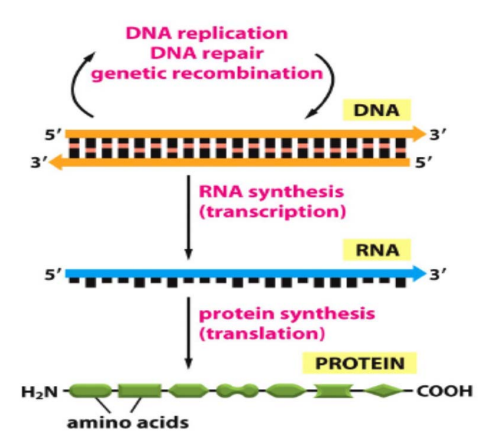
Mathematical analysis of gene regulation activator model
This paper presents a complete analysis of the mathematical model of the gene regulation process. The model describes the induced gene expression under the effect of activators. The model differential equations are solved analytically, and the exact solution of the gene model is introduced. Moreover, a study of the model dynamics, including the fixed points and stability conditions are presented. The parameters effects on the phase plane portraits and the transient responses of the mRNA as well as the protein concentrations are intensively detailed. This work serves as a brick stone towards a
Comparative study of fractional filters for Alzheimer disease detection on MRI images
This paper presents a comparative study of four fractional order filters used for edge detection. The noise performance of these filters is analyzed upon the addition of random Gaussian noise, as well as the addition of salt and pepper noise. The peak signal to noise ratio (PSNR) of the detected images is numerically compared. The mean square error (MSE) of the detected images as well as the execution time are also adopted as evaluation methods for comparison. The visual comparison of the filters capability in medical image edge detection is presented, that can help in the diagnosis of
Implementation of a Pulsed-Wave Spectral Doppler Module on a Programmable Ultrasound System
Pulsed wave Doppler ultrasound is commonly used in the diagnosis of cardiovascular and blood flow abnormalities. Doppler techniques have gained clinical significance due to its safety, real-time performance and affordability. This work presents the development of a pulsed wave spectral Doppler module, which was integrated into a reconfigurable ultrasound system. The targeted system adopts a hardware-software partitioning scheme where an FPGA handles the front-end and a PC performs the back-end. Two factors were considered during the design. First, the data transfer rate between hardware and
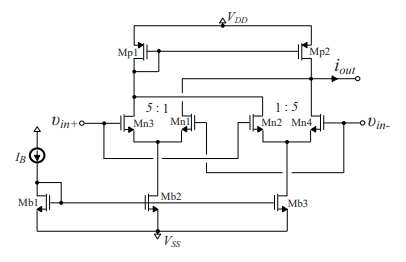
Design of fractional-order differentiator-lowpass filters for extracting the R peaks in ECG signals
An implementation of a fractional-order differentiator-lowpass filter is presented in this work, which is constructed from Operational Transconductance Amplifiers as active cells. This offers the benefits of electronic tuning and, also, of monolithic implementation. The presented scheme has been employed for the extraction of the R peaks in electrocardiogram signals due to its efficiency for performing this task even in a noisy environment. The provided post-layout simulation results confirm the correct operation of this solution as well as its reasonable sensitivity characteristics. © 2019
Pagination
- Previous page ‹‹
- Page 5
- Next page ››