Detection of cardiac function abnormality from MRI images using normalized wall thickness temporal patterns
Purpose. To develop a method for identifying abnormal myocardial function based on studying the normalized wall motion pattern during the cardiac cycle. Methods. The temporal pattern of the normalized myocardial wall thickness is used as a feature vector to assess the cardiac wall motion abnormality. Principal component analysis is used to reduce the feature dimensionality and the maximum likelihood method is used to differentiate between normal and abnormal features. The proposed method was applied on a dataset of 27 cases from normal subjects and patients. Results. The developed method
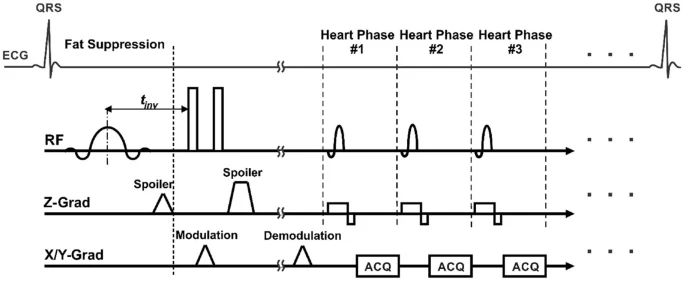
An efficient fat suppression technique for stimulated-echo based CMR
[No abstract available]
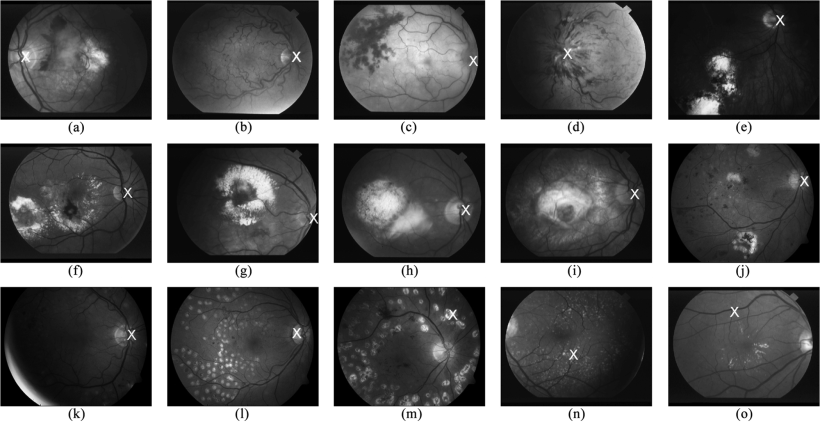
Fast localization of the optic disc using projection of image features
Optic Disc (OD) localization is an important pre-processing step that significantly simplifies subsequent segmentation of the OD and other retinal structures. Current OD localization techniques suffer from impractically-high computation times (few minutes per image). In this work, we present a fast technique that requires less than a second to localize the OD. The technique is based upon obtaining two projections of certain image features that encode the x- and y- coordinates of the OD. The resulting 1-D projections are then searched to determine the location of the OD. This avoids searching
CoCoNUT: An efficient system for the comparison and analysis of genomes
Background: Comparative genomics is the analysis and comparison of genomes from different species. This area of research is driven by the large number of sequenced genomes and heavily relies on efficient algorithms and software to perform pairwise and multiple genome comparisons. Results: Most of the software tools available are tailored for one specific task. In contrast, we have developed a novel system CoCoNUT (Computational Comparative geNomics Utility Toolkit) that allows solving several different tasks in a unified framework: (1) finding regions of high similarity among multiple genomic
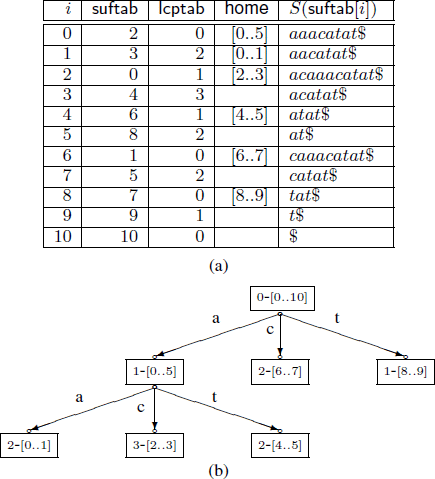
Fine tuning the enhanced suffix array
The enhanced suffix array is an indexing data structure used for a wide range of applications in Bioinformatics. It is basically the suffix array but enhanced with extra tables that provide extra information to improve the performance in theory and in practice. In this paper, we present a number of improvements to the enhanced suffix array: 1) We show how to find a pattern of length m in O(m) time, i.e., independent of the alphabet size. 2) We present an improved representation of the bucket table. 3) We improve the access time of addressing the LCP (longest common prefix) table when one byte
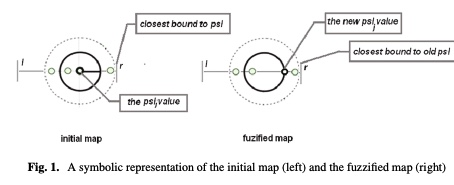
A fuzzy approach of sensitivity for multiple colonies on ant colony optimization
In order to solve combinatorial optimization problem are used mainly hybrid heuristics. Inspired from nature, both genetic and ant colony algorithms could be used in a hybrid model by using their benefits. The paper introduces a new model of Ant Colony Optimization using multiple colonies with different level of sensitivity to the ant’s pheromone. The colonies react different to the changing environment, based on their level of sensitivity and thus the exploration of the solution space is extended. Several discussion follows about the fuzziness degree of sensitivity and its influence on the
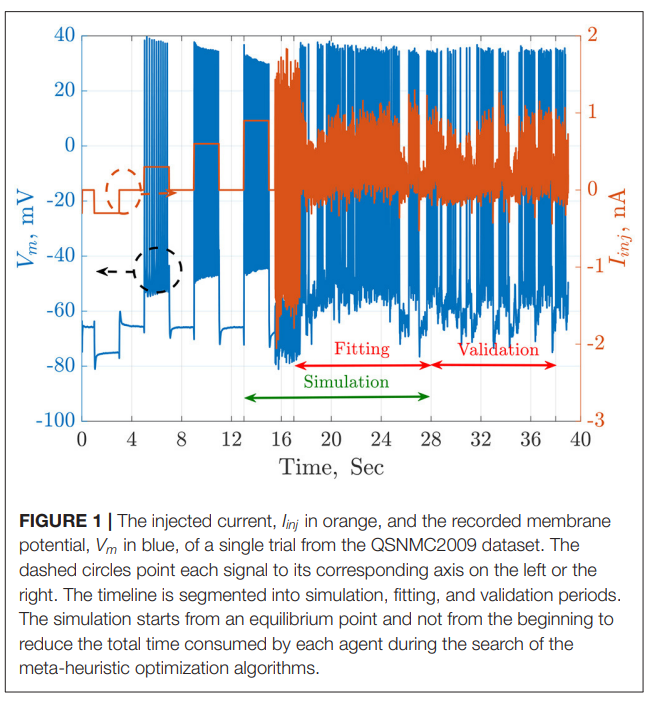
Parameter Estimation of Two Spiking Neuron Models With Meta-Heuristic Optimization Algorithms
The automatic fitting of spiking neuron models to experimental data is a challenging problem. The integrate and fire model and Hodgkin–Huxley (HH) models represent the two complexity extremes of spiking neural models. Between these two extremes lies two and three differential-equation-based models. In this work, we investigate the problem of parameter estimation of two simple neuron models with a sharp reset in order to fit the spike timing of electro-physiological recordings based on two problem formulations. Five optimization algorithms are investigated; three of them have not been used to
Constructing suffix array during decompression
The suffix array is an indexing data structure used in a wide range of applications in Bioinformatics. Biological DNA sequences are available to download from public servers in the form of compressed files, where the popular lossless compression program gzip [1] is employed. The straightforward method to construct the suffix array for this data involves decompressing the sequence file, storing it on disk, and then calling a suffix array construction program to build the suffix array. This scenario, albeit feasible, requires disk access and throws away valuable information in the compressed
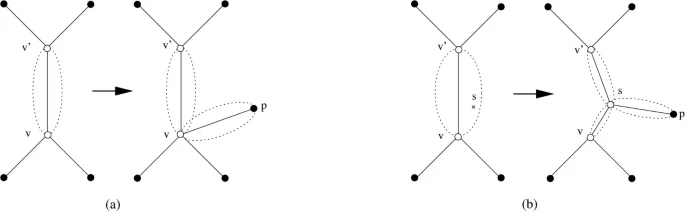
A fast algorithm for the multiple genome rearrangement problem with weighted reversals and transpositions
Background: Due to recent progress in genome sequencing, more and more data for phylogenetic reconstruction based on rearrangement distances between genomes become available. However, this phylogenetic reconstruction is a very challenging task. For the most simple distance measures (the breakpoint distance and the reversal distance), the problem is NP-hard even if one considers only three genomes. Results: In this paper, we present a new heuristic algorithm that directly constructs a phylogenetic tree w.r.t. the weighted reversal and transposition distance. Experimental results on previously
Pagination
- Previous page ‹‹
- Page 2
- Next page ››