AmpliconNet: Sequence Based Multi-layer Perceptron for Amplicon Read Classification Using Real-time Data Augmentation
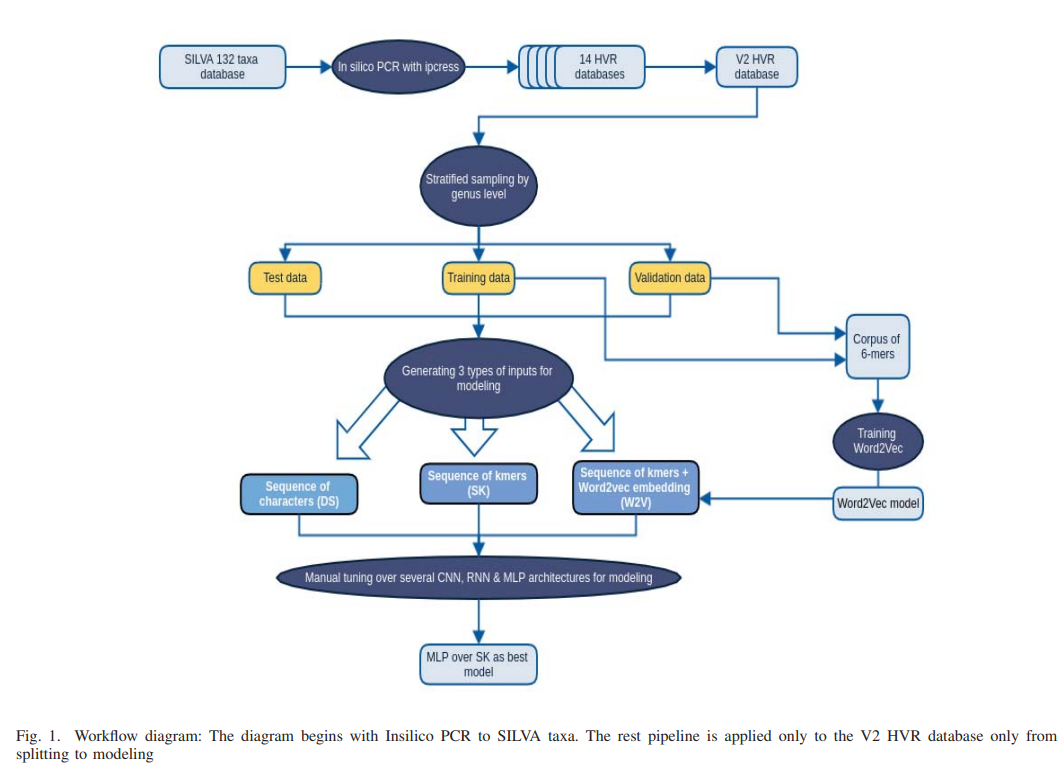
Taxonomic assignment is the core of targeted metagenomics approaches that aims to assign sequencing reads to their corresponding taxonomy. Sequence similarity searching and machine learning (ML) are two commonly used approaches for taxonomic assignment based on the 16S rRNA. Similarity based approaches require high computation resources, while ML approaches dont need these resources in prediction. The majority of these ML approaches depend on k-mer frequency rather than direct sequence, which leads to low accuracy on short reads as k-mer frequency doesnt consider k-mer position. Moreover training ML taxonomic classifiers depend on a specific read length which may reduce the prediction performance by decreasing read length. In this study, we built a neural network classifier for 16S rRNA reads based on SILVA database (version 132). Modeling was performed on direct sequences using Convolutional neural network (CNN) and other neural network architectures such as Multi-layer Perceptron and Recurrent Neural Network. In order to reduce modeling time of the direct sequences, In-silico PCR was applied on SILVA database. Total number of 14 subset databases were generated by universal primers for each single or paired high variable region (HVR). Moreover, in this study, we illustrate the results for the V2 database model on 1850 classes on the genus level. In order to simulate sequencing fragmentation, we trained variable length subsequences from 50 bases till the full length of the HVR that are randomly changing in each training iteration. Simple MLP model with global max pooling gives 0.93 test accuracy for the genus level (for reads of 100 base subsequences) and 0.96 accuracy for the genus level respectively (on the full length V2 HVR). In this study, we present a novel method AmpliconNet https://github.com/ali-kishk/AmpliconNet to model the direct amplicon sequence using MLP over a sequence of k-mers faster 20 times than CNN in training and 10 times in prediction. © 2018 IEEE.