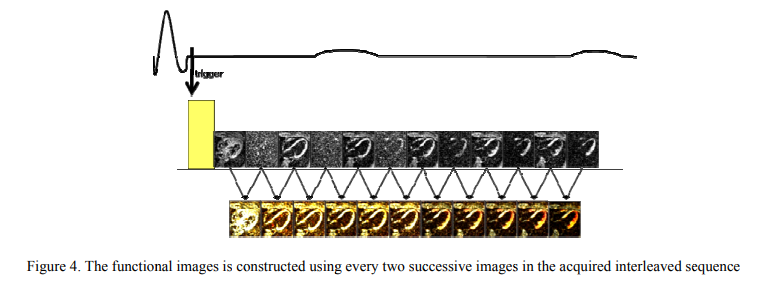
Strain correction in interleaved strain-encoded (SENC) cardiac MR
The strain encoding (SENC) technique directly encodes regional strain of the heart into the acquired MR images and produces two images with two different tunings so that longitudinal strain, on the short-axis view, or circumferential strain on the long-axis view, are measured. Interleaving acquisition is used to shorten the acquisition time of the two tuned images by 50%, but it suffers from errors in the strain calculations due to inter-tunings motion of the heart. In this work, we propose a method to correct for the inter-tunings motion by estimating the motion-induced shift in the spatial
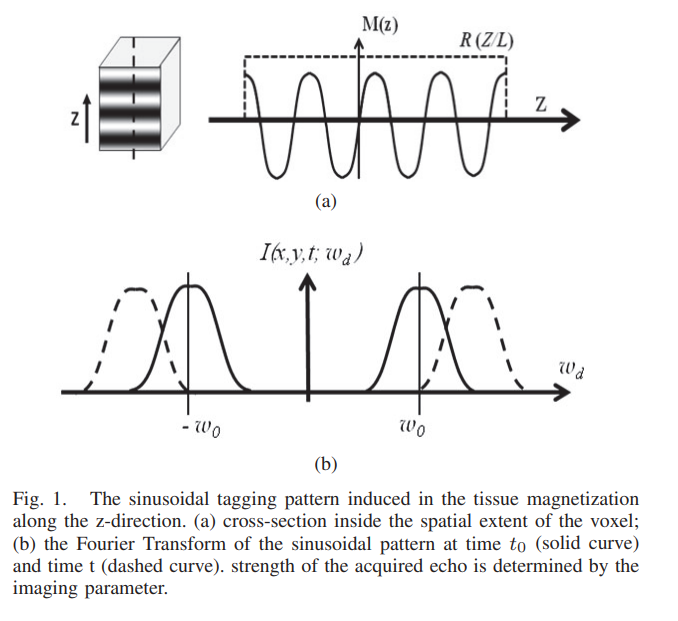
Maximum likelihood estimator for signal intensity in STEAM-based MR imaging techniques
Stimulated echo acquisition mode (STEAM) is a generic imaging technique that lies at the core of many magnetic resonance imaging (MRI) techniques such MRI tagging, displacement encoded MRI, black-blood cardiac imaging. Nevertheless, tissue deformation causes frequency shift of the MR signal and leads to severe signal attenuation. In this work, a maximum likelihood estimator for the signal amplitude is proposed and used to correct the image artifacts. Numerical simulation and real MR data are used to test and validate the proposed method. © 2011 IEEE.
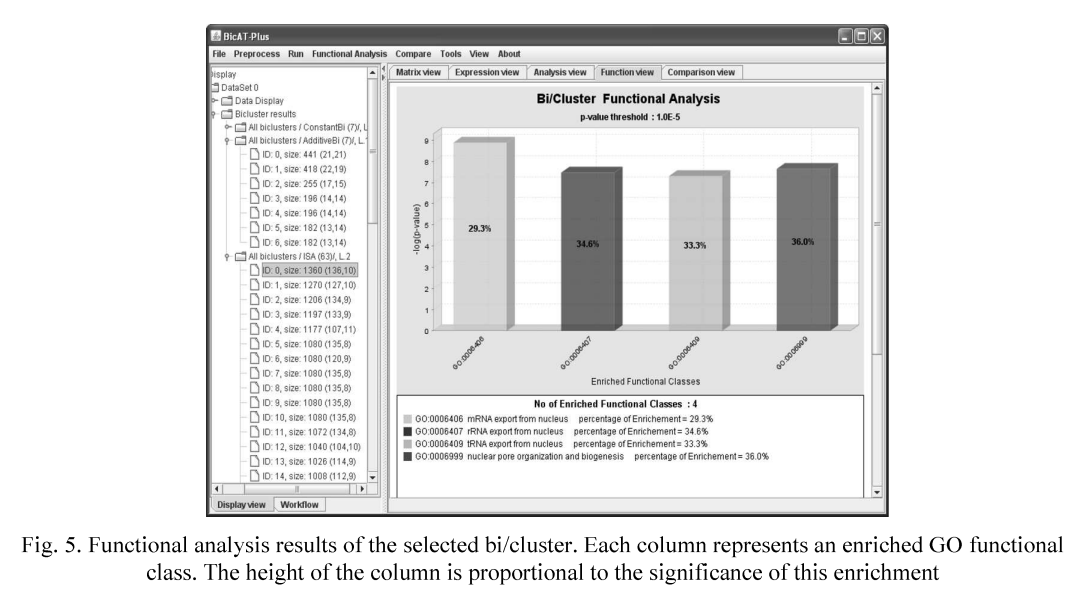
BicATPlus: An automatic comparative tool for Bi/Clustering of gene expression data obtained using microarrays
In the last few years the gene expression microarray technology has become a central tool in the field of functional genomics in which the expression levels of thousands of genes in a biological sample are determined in a single experiment. Several clustering and biclustering methods have been introduced to analyze the gene expression data by identifying the similar patterns and grouping genes into subsets that share biological significance. However, it is not clear how the different methods compare with each other with respect to the biological relevance of the biclusters and clusters as well
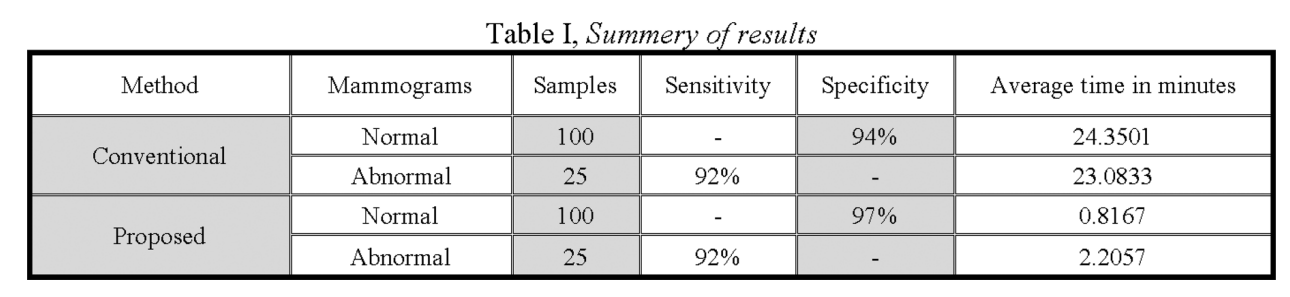
Fast fractal modeling of mammograms for microcalcifications detection
Clusters of microcalcifications in mammograms are an important early sign of breast cancer in women. Comparing with microcalcifications, the breast background tissues have high local self-similarity, which is the basic property of fractal objects. A fast fractal modeling method of mammograms for detecting the presence of microcalcifications is proposed in this paper. The conventional fractal modeling method consumes too much computation time. In the proposed method, the image is divided into shade (homogeneous) and non-shade blocks based on the dynamic range and only the non-shade blocks are
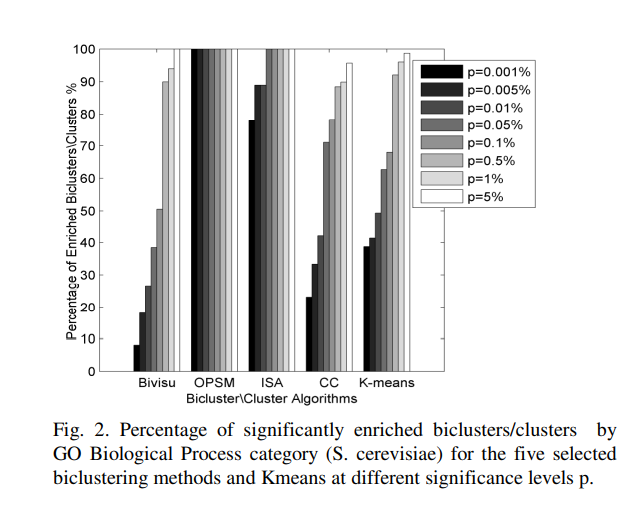
An automatic gene ontology software tool for bicluster and cluster comparisons
We propose an Automatic Gene Ontology (AGO) software as a flexible, open-source Matlab software tool that allows the user to easily compare the results of the bicluster and cluster methods. This software provides several methods to differentiate and compare the results of candidate algorithms. The results reveal that bicluster/cluster algorithms could be considered as integrated modules to recover the interesting patterns in the microarray datasets. The further application of AGO could to solve the dimensionality reduction of the gene regulatory networks. Availability: AGO and help file is
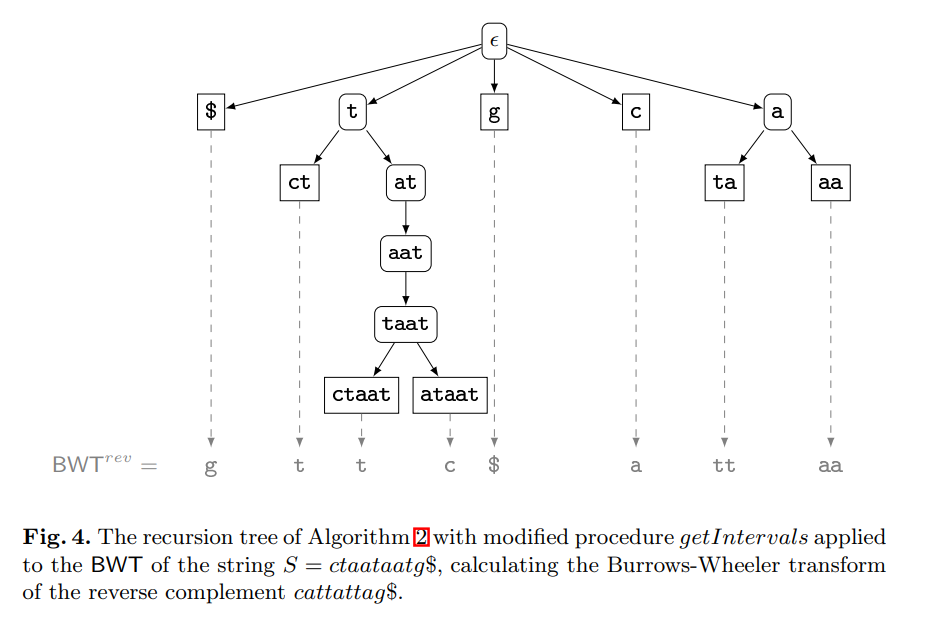
Computing the burrows-wheeler transform of a string and its reverse
The contribution of this paper is twofold. First, we provide new theoretical insights into the relationship between a string and its reverse: If the Burrows-Wheeler transform (BWT) of a string has been computed by sorting its suffixes, then the BWT and the longest common prefix array of the reverse string can be derived from it without suffix sorting. Furthermore, we show that the longest common prefix arrays of a string and its reverse are permutations of each other. Second, we provide a parallel algorithm that, given the BWT of a string, computes the BWT of its reverse much faster than all
Developing a Greenometer for green manufacturing assessment
In this paper a toolbox (Greenometer) to assess the greenness level of manufacturing companies is proposed. The assessment approach is based on capturing the relative greenness position of any company among other industries from different sectors as well as within the same sector. The assessment was based on selected greenness attributes and their composing indicators at each of the two levels of the developed Greenometer. Geometric Mean Method (GMM) was adopted to be the generic assessment technique for cross industries greenness evaluation, while Data Envelopment Analysis (DEA) was employed
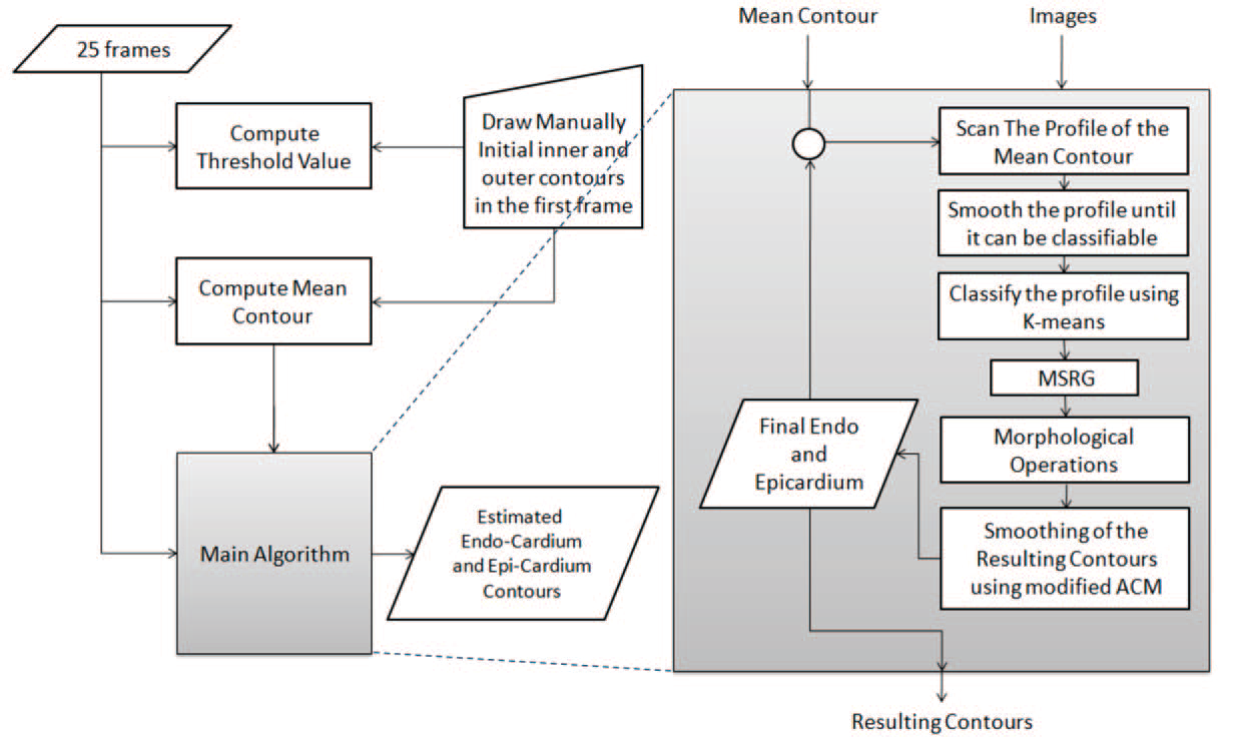
Segmentation of left ventricle in cardiac MRI images using adaptive multi-seeded region growing
Multi-slice short-axis acquisitions of the left ventricle are fundamental for estimating the volume and mass of the left ventricle in cardiac MRI scans. Manual segmentation of the myocardium in all time frames per each cross-section is a cumbersome task. Therefore, automatic myocardium segmentation methods are essential for cardiac functional analysis. Region growing has been proposed to segment the myocardium. Although the technique is simple and fast, non uniform intensity and low-contrast interfaces of the myocardium are major challenges of the technique that limit its use in myocardial
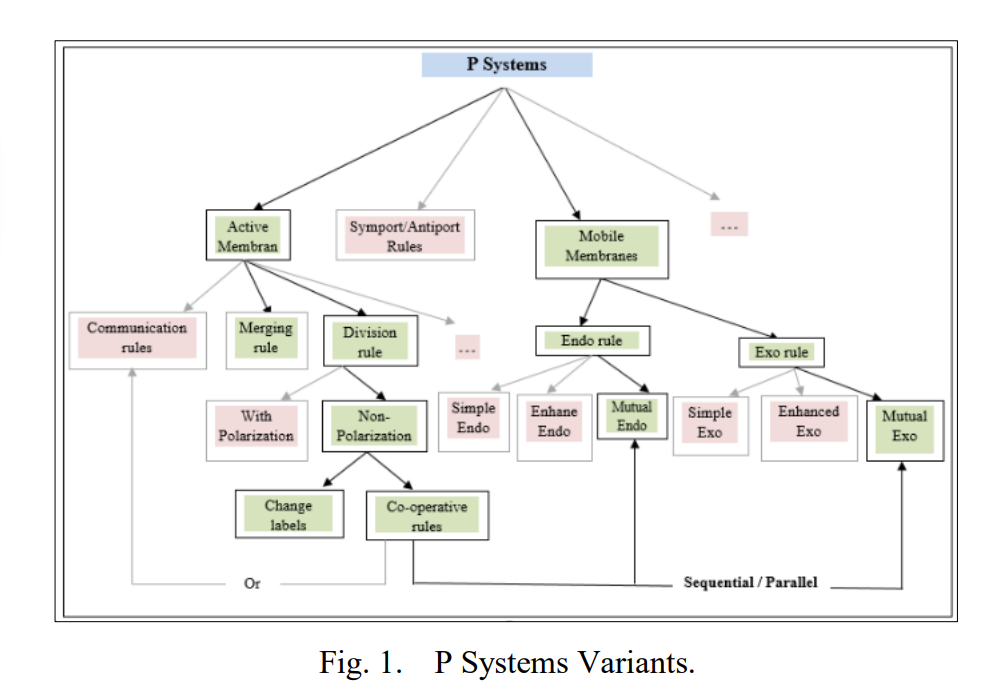
P Systems Implementation: A Model of Computing for Biological Mitochondrial Rules using Object Oriented Programming
Membrane computing is a computational framework that depends on the behavior and structure of living cells. P systems are arising from the biological processes which occur in the living cells’ organelles in a non-deterministic and maximally parallel manner. This paper aims to build a powerful computational model that combines the rules of active and mobile membranes, called Mutual Dynamic Membranes (MDM). The proposed model will describe the biological mechanisms of the metabolic regulation of mitochondrial dynamics made by mitochondrial membranes. The behaviors of the proposed model regulate
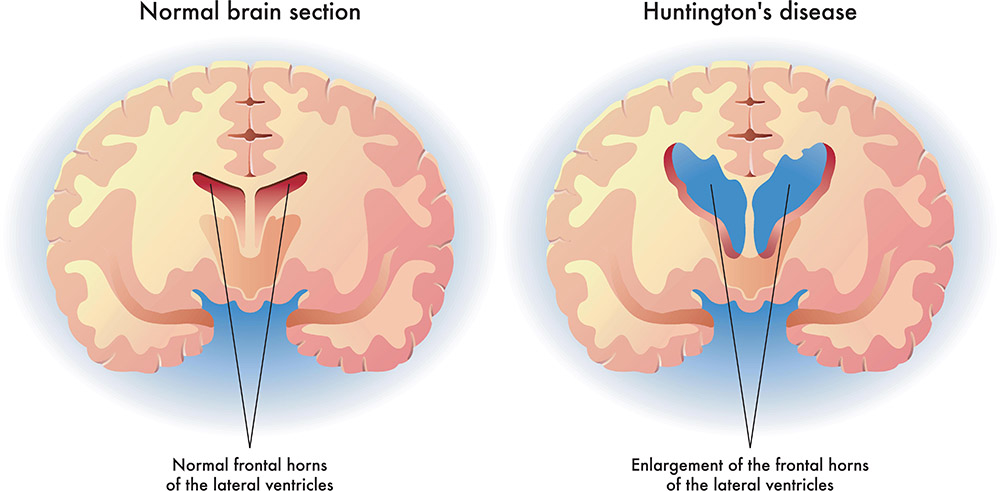
INVESTIGATION OF DIFFERENTIALLY EXPRESSED GENE RELATED TO HUNTINGTON'S DISEASE USING GENETIC ALGORITHM
neurodegenerative diseases have complex pathological mechanisms. Detecting disease-associated genes with typical differentially expressed gene selection approaches are ineffective. Recent studies have shown that wrappers Evolutionary optimization methods perform well in feature selection for high dimensional data, but they are computationally costly. This paper proposes a simple method based on a genetic algorithm engaged with the Empirical Bays T-statistics test to enhance the disease-associated gene selection process. The proposed method is applied to Affymetrix microarray data from
Pagination
- Previous page ‹‹
- Page 16
- Next page ››